MSH2 (mutS homolog 2)
- symbol:
- MSH2
- locus group:
- protein-coding gene
- location:
- 2p21-p16.3
- gene_family:
- alias symbol:
- HNPCC|HNPCC1|MSH-2
- alias name:
- DNA mismatch repair protein Msh2
- entrez id:
- 4436
- ensembl gene id:
- ENSG00000095002
- ucsc gene id:
- uc002rvy.3
- refseq accession:
- NM_000251
- hgnc_id:
- HGNC:7325
- approved reserved:
- 1993-07-28
MSH2(MutS homolog 2)是DNA错配修复(MMR)系统中的关键基因,属于MutS基因家族。该基因家族成员在进化上高度保守,主要功能是识别和修复DNA复制过程中产生的碱基错配或插入缺失错误,维持基因组稳定性。MSH2蛋白与MSH6或MSH3结合形成MutSα或MutSβ复合物,其中MutSα负责识别单碱基错配和小插入缺失,而MutSβ主要处理较大的插入缺失错误。MSH2基因突变会导致错配修复功能缺陷,造成微卫星不稳定(MSI),这是林奇综合征(遗传性非息肉病性结直肠癌)的主要病因。约40-50%的林奇综合征病例由MSH2突变引起,患者易患结直肠癌、子宫内膜癌等多种恶性肿瘤。MSH2突变还可能导致Turcot综合征和 Muir-Torre综合征。当MSH2表达降低时,细胞无法有效修复DNA复制错误,突变累积加速,促进肿瘤发生。过表达MSH2虽不直接致病,但可能干扰其他修复蛋白的平衡。MSH2还与化疗耐药相关,其缺失可能导致对某些化疗药物(如5-氟尿嘧啶)敏感性降低。该基因位于人类染色体2p21-p16区域,包含16个外显子。MutS家族成员均含有保守的ATP酶结构域和错配结合域,通过ATP依赖的方式启动修复过程。除MSH2外,家族还包括MSH3、MSH6等,它们在功能上既分工又协作,共同维护基因组完整性。检测MSH2突变和微卫星不稳定状态对林奇综合征筛查和肿瘤个体化治疗具有重要意义。
This locus is frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). When cloned, it was discovered to be a human homolog of the E. coli mismatch repair gene mutS, consistent with the characteristic alterations in microsatellite sequences (RER+ phenotype) found in HNPCC. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2012]
这个位点突变频率在遗传性非息肉结肠癌(HNPCC)。当克隆时,被发现是大肠杆菌错配修复基因mutS的的人同源物,与微卫星序列中HNPCC发现的特征的改变(RER +表型)相一致。已发现该基因编码不同亚型的两个转录变异体。 [由RefSeq的,2012年4月提供]
基因本体信息
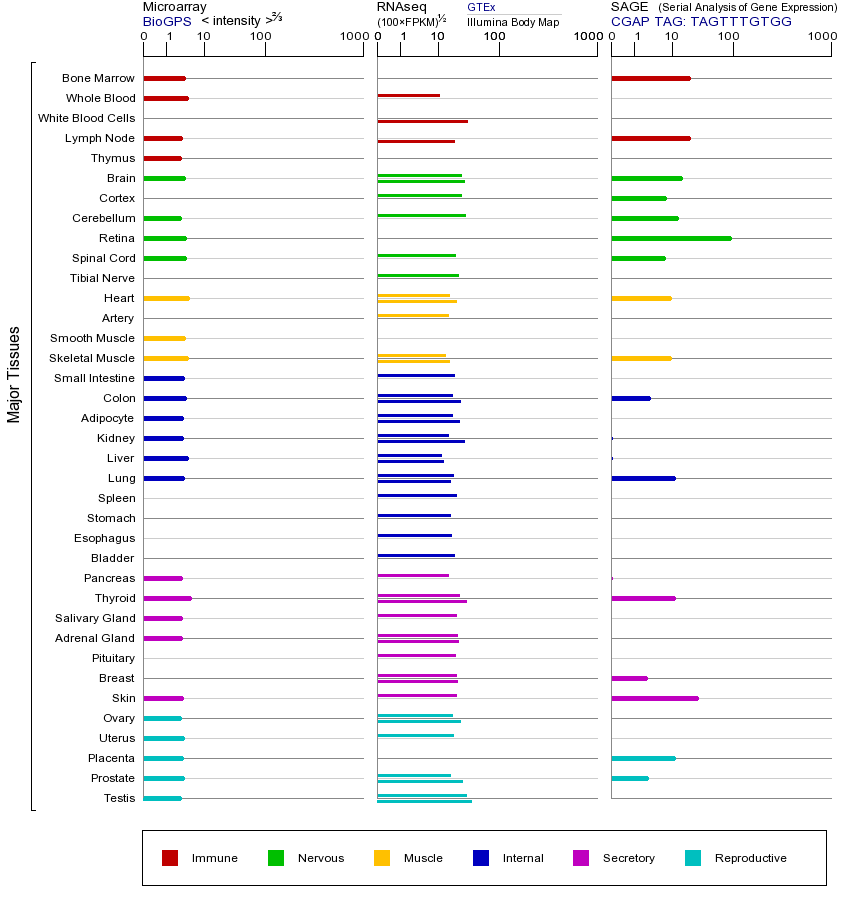
MSH2基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
MSH2基因的本体(GO)信息:
| 名称 |
|---|
| 3430 Mismatch repair [PATH:hsa03430] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5210 Colorectal cancer [PATH:hsa05210] |
| 名称 |
|---|
| DNA Repair |
| Mismatch Repair |
| Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) |
| Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Torre-Muir syndrome | 0.375220608 | 36 | 2 | BeFree_CLINVAR_CTD_human_LHGDN_ORPHANET |
| Turcot syndrome (disorder) | 0.365157396 | 19 | 6 | BeFree_CLINVAR_CTD_human_ORPHANET |
| Hereditary Nonpolyposis Colorectal Cancer | 0.33183516 | 428 | 423 | BeFree_CLINVAR_GAD_ORPHANET |
| Colorectal cancer, hereditary nonpolyposis, type 1 | 0.32 | 30 | 27 | CLINVAR_MGD_UNIPROT |
| Hereditary Nonpolyposis Colorectal Neoplasms | 0.244973608 | 54 | 0 | CTD_human_GAD_LHGDN |
| Colorectal Neoplasms | 0.201143185 | 41 | 0 | BeFree_CTD_human_GAD_LHGDN |
| Colorectal Cancer | 0.14378884 | 287 | 4 | BeFree_GAD |
| Glioma | 0.123181358 | 4 | 0 | BeFree_CTD_human_GAD |
| Neoplastic Syndromes, Hereditary | 0.121357209 | 5 | 58 | BeFree_CLINVAR |
| Neurofibromatosis 1 | 0.121085767 | 4 | 0 | BeFree_CTD_human |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。