MLH1 (mutL homolog 1)
- symbol:
- MLH1
- locus group:
- protein-coding gene
- location:
- 3p22.2
- gene_family:
- alias symbol:
- HNPCC|FCC2|HNPCC2|MLH-1
- alias name:
- None
- entrez id:
- 4292
- ensembl gene id:
- ENSG00000076242
- ucsc gene id:
- uc003cgl.4
- refseq accession:
- NM_000249
- hgnc_id:
- HGNC:7127
- approved reserved:
- 1993-11-24
MLH1(MutL homolog 1)是DNA错配修复(MMR)系统中的关键基因,属于MutL基因家族。该家族成员在DNA复制过程中负责识别和修复碱基错配,维持基因组稳定性。MLH1基因编码的蛋白质与其他MMR蛋白(如MSH2、MSH6)形成复合物,共同参与修复DNA复制错误。MLH1蛋白的主要作用位点是细胞核,通过与PMS2结合形成MutLα复合物,在修复过程中发挥核心作用。MLH1突变会导致错配修复功能缺陷,造成微卫星不稳定性(MSI),进而增加癌症风险,尤其是林奇综合征(遗传性非息肉病性结直肠癌,HNPCC)的主要致病基因之一。MLH1突变携带者患结直肠癌、子宫内膜癌、卵巢癌等恶性肿瘤的风险显著升高。MLH1表达降低或功能丧失会导致细胞无法有效修复DNA复制错误,积累突变并促进肿瘤发生。MLH1过表达的情况较少见,但可能通过过度修复干扰正常DNA代谢。MLH1的表观遗传沉默(如启动子甲基化)是散发性结直肠癌的常见机制。MLH1基因家族(MutL家族)还包括MLH3、PMS1和PMS2等成员,它们均参与DNA错配修复,但功能有所分化。该家族蛋白均含有保守的ATP酶结构域和蛋白质相互作用域,能够识别错配DNA并招募其他修复因子。MLH1功能异常不仅影响基因组稳定性,还可能改变细胞对化疗药物的敏感性,例如MLH1缺陷肿瘤可能对某些DNA损伤药物(如铂类)更敏感,但对5-氟尿嘧啶耐药。研究MLH1有助于理解癌症发生机制并指导个性化治疗策略。
This gene was identified as a locus frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). It is a human homolog of the E. coli DNA mismatch repair gene mutL, consistent with the characteristic alterations in microsatellite sequences (RER+phenotype) found in HNPCC. Alternative splicing results in multiple transcript variants encoding distinct isoforms. Additional transcript variants have been described, but their full-length natures have not been determined.[provided by RefSeq, Nov 2009]
该基??因被鉴定为在遗传性非息肉结肠直肠癌(HNPCC)频繁突变的轨迹。它是大肠杆菌DNA错配修复基因mutL的人类同源,与微卫星序列中发现HNPCC的特点改变(RER +表型)一致。在多个转录剪接变异体结果不同编码的亚型。额外转录变体进行了说明,但它们的全长性质尚未确定。[通过的RefSeq,2009年11月提供]
基因本体信息
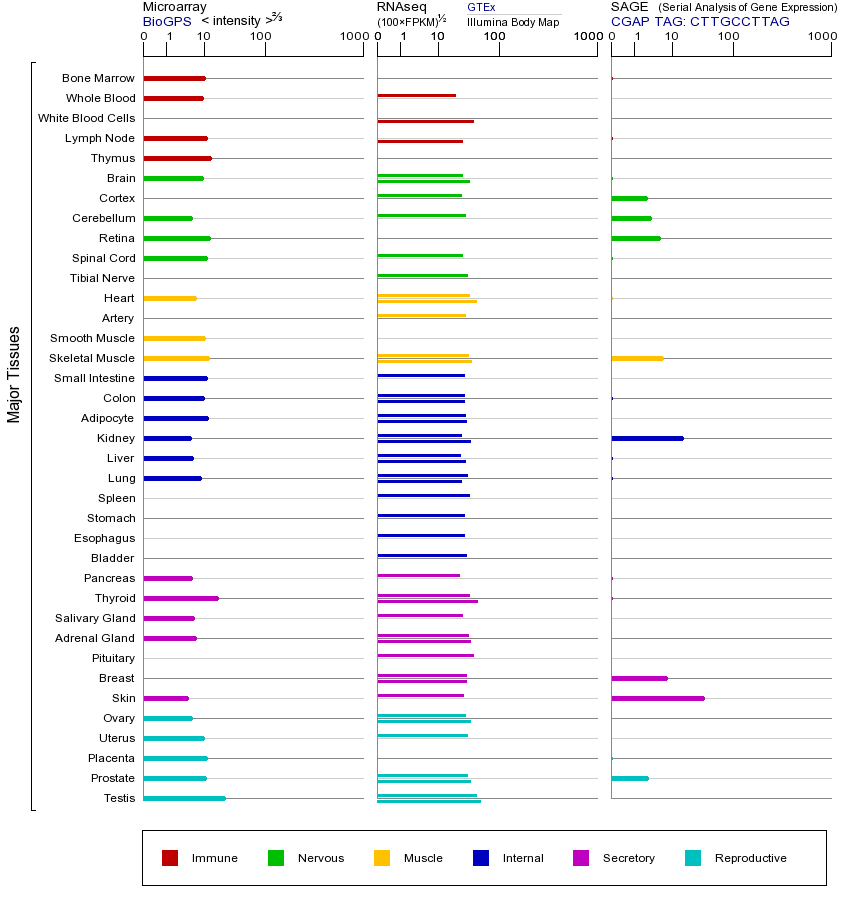
MLH1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
MLH1基因的本体(GO)信息:
| 名称 |
|---|
| 3430 Mismatch repair [PATH:hsa03430] |
| 3460 Fanconi anemia pathway [PATH:hsa03460] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5210 Colorectal cancer [PATH:hsa05210] |
| 5213 Endometrial cancer [PATH:hsa05213] |
| 名称 |
|---|
| Cell Cycle |
| DNA Repair |
| Meiosis |
| Meiotic recombination |
| Mismatch Repair |
| Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) |
| Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Turcot syndrome (disorder) | 0.492486326 | 47 | 7 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Hereditary Non-Polyposis Colon Cancer Type 2 | 0.440271442 | 39 | 18 | BeFree_CLINVAR_CTD_human_MGD_UNIPROT |
| Torre-Muir syndrome | 0.365971721 | 22 | 0 | BeFree_CLINVAR_CTD_human_ORPHANET |
| Hereditary Nonpolyposis Colorectal Cancer | 0.329468128 | 439 | 472 | BeFree_CLINVAR_GAD_ORPHANET |
| Hereditary Nonpolyposis Colorectal Neoplasms | 0.239826141 | 52 | 0 | CTD_human_GAD_LHGDN |
| Colorectal Neoplasms | 0.23010898 | 72 | 0 | BeFree_CTD_human_GAD_LHGDN |
| Colorectal Cancer | 0.16 | 422 | 19 | BeFree_GAD |
| ovarian neoplasm | 0.133068936 | 12 | 0 | BeFree_CTD_human_LHGDN |
| Prostatic Neoplasms | 0.1254487 | 3 | 0 | CTD_human_LHGDN |
| Neoplasm Metastasis | 0.125438769 | 12 | 0 | BeFree_CTD_human_LHGDN |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。