ITGAV (integrin subunit alpha V)
- symbol:
- ITGAV
- locus group:
- protein-coding gene
- location:
- 2q32.1
- gene_family:
- CD molecules|Integrins
- alias symbol:
- CD51
- alias name:
- None
- entrez id:
- 3685
- ensembl gene id:
- ENSG00000138448
- ucsc gene id:
- uc002upq.5
- refseq accession:
- NM_002210
- hgnc_id:
- HGNC:6150
- approved reserved:
- 1988-07-19
ITGAV基因编码整合素αV亚基,属于整合素超家族中的αV亚家族。整合素是由α和β亚基组成的异源二聚体跨膜受体,主要介导细胞与细胞外基质(ECM)的黏附以及细胞间信号传导。ITGAV可与多种β亚基(如β1、β3、β5、β6、β8)结合形成不同的整合素受体,其中αVβ3和αVβ5最为常见。这些整合素能识别含有RGD(精氨酸-甘氨酸-天冬氨酸)序列的ECM蛋白,如纤连蛋白、玻连蛋白、骨桥蛋白等,参与细胞迁移、增殖、存活和分化等过程。ITGAV在胚胎发育、血管生成、伤口愈合、骨重塑和免疫应答中发挥关键作用。该基因突变可能导致多种疾病,如ITGAV突变与Glanzmann血小板无力症相关,患者表现出出血倾向;αVβ3和αVβ5的异常表达与肿瘤转移、骨质疏松和纤维化疾病密切相关。ITGAV过表达常见于多种癌症(如乳腺癌、胶质母细胞瘤),促进肿瘤细胞侵袭转移和血管生成;而敲除ITGAV会导致小鼠胚胎致死,显示其在发育中的必要性。ITGAV还参与TGF-β的激活,其表达降低可能影响组织纤维化和免疫调节。ITGAV属于整合素α亚基家族,该家族成员均含有7个保守的N端重复序列,其中4个含有二价阳离子结合位点,对配体识别至关重要。目前针对ITGAV的靶向药物(如西仑吉肽)正在研发中,用于抑制肿瘤血管生成。
This gene encodes a protein that is a member of the integrin superfamily. Integrins are heterodimeric integral membrane proteins composed of an alpha chain and a beta chain. This protein undergoes post-translational cleavage to yield disulfide-linked heavy and light chains that combine with multiple integrin beta chains to form different integrins. This protein has been shown to heterodimerize with beta 1, beta 3, beta 5, beta 6, and beta 8; the heterodimer of alpha v and beta 3 is the Vitronectin receptor. This protein interacts with several extracellular matrix proteins to mediate cell adhesion and may play a role in cell migration. It is proposed that this protein may regulate angiogenesis and cancer progression. Alternative splicing results in multiple transcript variants that encode different protein isoforms. Note that the integrin alpha 5 and integrin alpha V chains are produced by distinct genes. [provided by RefSeq, Jan 2015]
这个基因编码的蛋白质,它是整联超家族的成员。整合素是一个α链和一个β链组成的异二聚体整合膜蛋白。这种蛋白质经历翻译后切割产生二硫键连接的重链和轻链是具有多个整β链结合以形成不同的整联。这种蛋白质已被证实与β-1,β3,β5,β6和β8异源二聚体;阿尔法V和β3的异源二聚体是玻连蛋白受体。这种蛋白质与几个细胞外基质蛋白介导细胞粘附相互作用并可能在细胞的迁移的作用。有人建议,这种蛋白质可调节血管生成和癌症的发展。选择性剪接结果在多个抄本变形编码不同的蛋白质亚型。需要注意的是整合素5和整合素V链由不同的基因产生的。 [由RefSeq的,2015年1月提供]
基因本体信息
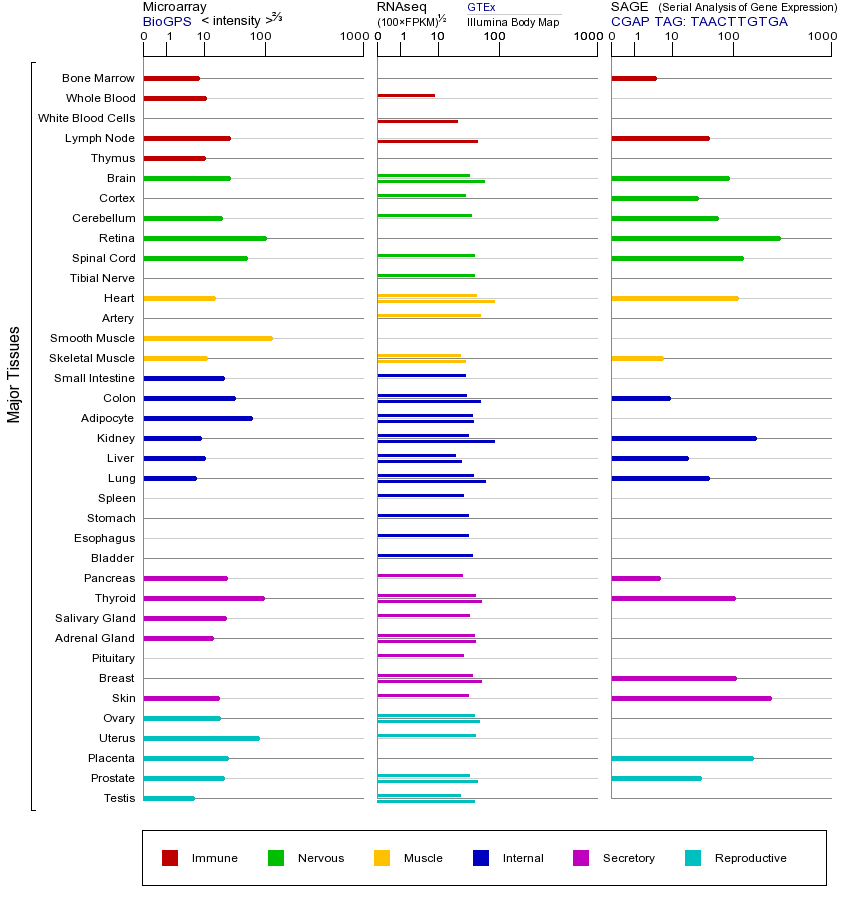
ITGAV基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
ITGAV基因的本体(GO)信息:
| 名称 |
|---|
| 4151 PI3K-Akt signaling pathway [PATH:hsa04151] |
| 4512 ECM-receptor interaction [PATH:hsa04512] |
| 4514 Cell adhesion molecules (CAMs) [PATH:hsa04514] |
| 4145 Phagosome [PATH:hsa04145] |
| 4810 Regulation of actin cytoskeleton [PATH:hsa04810] |
| 4510 Focal adhesion [PATH:hsa04510] |
| 4919 Thyroid hormone signaling pathway [PATH:hsa04919] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5205 Proteoglycans in cancer [PATH:hsa05205] |
| 5222 Small cell lung cancer [PATH:hsa05222] |
| 5410 Hypertrophic cardiomyopathy (HCM) [PATH:hsa05410] |
| 5412 Arrhythmogenic right ventricular cardiomyopathy (ARVC) [PATH:hsa05412] |
| 5414 Dilated cardiomyopathy (DCM) [PATH:hsa05414] |
| 名称 |
|---|
| Adaptive Immune System |
| Antigen processing-Cross presentation |
| Axon guidance |
| Cell surface interactions at the vascular wall |
| Class I MHC mediated antigen processing & presentation |
| Cross-presentation of particulate exogenous antigens (phagosomes) |
| Developmental Biology |
| ECM proteoglycans |
| Elastic fibre formation |
| Extracellular matrix organization |
| Hemostasis |
| Immune System |
| Integrin cell surface interactions |
| L1CAM interactions |
| Laminin interactions |
| Molecules associated with elastic fibres |
| Non-integrin membrane-ECM interactions |
| PECAM1 interactions |
| Signal Transduction |
| Signal transduction by L1 |
| Signaling by VEGF |
| Syndecan interactions |
| VEGFA-VEGFR2 Pathway |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Cerebrovascular accident | 0.12 | 1 | 0 | CTD_human |
| Cerebral Hemorrhage | 0.12 | 1 | 0 | CTD_human |
| Corneal Neovascularization | 0.08 | 1 | 0 | RGD |
| Ischemia | 0.08 | 1 | 0 | RGD |
| Coronary Stenosis | 0.08 | 1 | 0 | RGD |
| Choroidal Neovascularization | 0.08 | 1 | 0 | RGD |
| Hypertensive disease | 0.08 | 1 | 0 | RGD |
| melanoma | 0.023413522 | 21 | 0 | BeFree_LHGDN |
| Mammary Neoplasms | 0.016346101 | 6 | 0 | LHGDN |
| Prostatic Neoplasms | 0.010897401 | 4 | 0 | LHGDN |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。