ERCC2 (ERCC excision repair 2, TFIIH core complex helicase subunit)
- symbol:
- ERCC2
- locus group:
- protein-coding gene
- location:
- 19q13.32
- gene_family:
- General transcription factors
- alias symbol:
- MAG|EM9|MGC102762|MGC126218|MGC126219
- alias name:
- excision repair cross-complementin…
- entrez id:
- 2068
- ensembl gene id:
- ENSG00000104884
- ucsc gene id:
- uc002pbj.3
- refseq accession:
- NM_000400
- hgnc_id:
- HGNC:3434
- approved reserved:
- 2001-06-22
ERCC2(Excision Repair Cross-Complementation Group 2)是一种关键的DNA修复基因,属于核苷酸切除修复(NER)通路中的核心成员。它编码的蛋白质是转录因子IIH(TFIIH)复合物的组成部分,该复合物在DNA损伤修复和转录调控中起重要作用。ERCC2的主要功能是通过其DNA解旋酶活性参与识别和修复由紫外线(UV)或化学物质引起的DNA损伤,特别是嘧啶二聚体等大体积损伤。该基因在维持基因组稳定性中至关重要,其突变会导致修复功能缺陷,引发严重的遗传病如着色性干皮病(XP)、科凯恩综合征(CS)或毛发硫营养不良(TTD),患者表现为对光敏感、早衰或神经发育异常。ERCC2属于ERCC基因家族,该家族成员均参与DNA修复,特别是NER通路,共同特点是能够识别并切除受损DNA片段,确保遗传信息完整性。若ERCC2表达降低,细胞会积累DNA损伤,增加突变率和癌症风险(如皮肤癌);而过度表达虽可能增强修复能力,但可能干扰正常细胞周期调控。ERCC2突变还与多种癌症易感性相关,例如肺癌和膀胱癌,其单核苷酸多态性(SNP)可作为生物标志物预测癌症风险或化疗反应。研究还发现ERCC2与其他修复基因(如XPA、XPC)协同作用,其功能异常会影响整个NER通路效率。
The nucleotide excision repair pathway is a mechanism to repair damage to DNA. The protein encoded by this gene is involved in transcription-coupled nucleotide excision repair and is an integral member of the basal transcription factor BTF2/TFIIH complex. The gene product has ATP-dependent DNA helicase activity and belongs to the RAD3/XPD subfamily of helicases. Defects in this gene can result in three different disorders, the cancer-prone syndrome xeroderma pigmentosum complementation group D, trichothiodystrophy, and Cockayne syndrome. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2008]
核苷酸切除修复途径是修复DNA损伤的机制。由该基因编码的蛋白参与转录偶联核苷酸切除修复和是基础转录因子BTF2 / TFIIH复杂的一个组成部件。该基因产物具有ATP依赖性的DNA解旋酶的活性和属于解旋酶的RAD3 / XPD亚科。在这个基因的缺陷可能导致三种不同的障碍,癌症倾向综合征着色性干皮互补组D,trichothiodystrophy和科凯恩综合征。已发现该基因编码不同亚型选择性剪接转录变异体。 [由RefSeq的,2008年8月提供]
基因本体信息
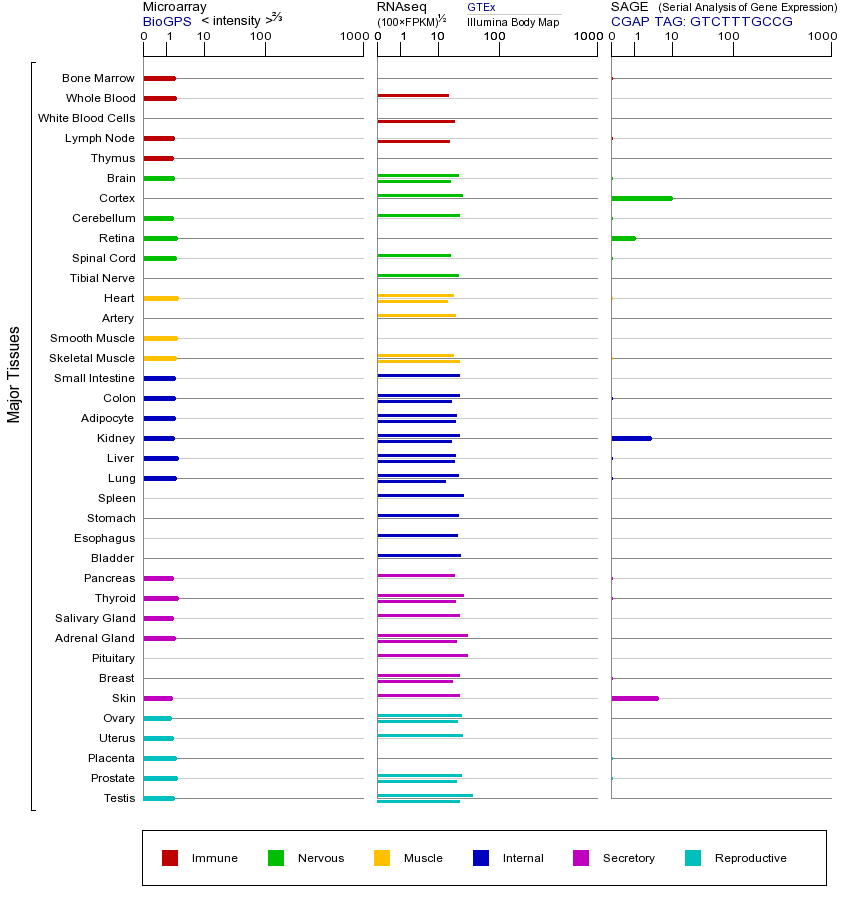
ERCC2基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
ERCC2基因的本体(GO)信息:
| 名称 |
|---|
| 3022 Basal transcription factors [PATH:hsa03022] |
| 3420 Nucleotide excision repair [PATH:hsa03420] |
| 名称 |
|---|
| Cytosolic iron-sulfur cluster assembly |
| Disease |
| DNA Repair |
| Dual incision reaction in GG-NER |
| Dual incision reaction in TC-NER |
| Epigenetic regulation of gene expression |
| Formation of HIV elongation complex in the absence of HIV Tat |
| Formation of HIV-1 elongation complex containing HIV-1 Tat |
| Formation of incision complex in GG-NER |
| Formation of RNA Pol II elongation complex |
| Formation of the Early Elongation Complex |
| Formation of the HIV-1 Early Elongation Complex |
| Formation of transcription-coupled NER (TC-NER) repair complex |
| Gene Expression |
| Global Genomic NER (GG-NER) |
| HIV Infection |
| HIV Life Cycle |
| HIV Transcription Elongation |
| HIV Transcription Initiation |
| Infectious disease |
| Late Phase of HIV Life Cycle |
| Metabolism |
| mRNA Capping |
| Negative epigenetic regulation of rRNA expression |
| NoRC negatively regulates rRNA expression |
| Nucleotide Excision Repair |
| RNA Pol II CTD phosphorylation and interaction with CE |
| RNA Polymerase I Chain Elongation |
| RNA Polymerase I Promoter Clearance |
| RNA Polymerase I Promoter Escape |
| RNA Polymerase I Transcription |
| RNA Polymerase I Transcription Initiation |
| RNA Polymerase I Transcription Termination |
| RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| RNA Polymerase II HIV Promoter Escape |
| RNA Polymerase II Pre-transcription Events |
| RNA Polymerase II Promoter Escape |
| RNA Polymerase II Transcription |
| RNA Polymerase II Transcription Elongation |
| RNA Polymerase II Transcription Initiation |
| RNA Polymerase II Transcription Initiation And Promoter Clearance |
| RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| Tat-mediated elongation of the HIV-1 transcript |
| Transcription |
| Transcription of the HIV genome |
| Transcription-coupled NER (TC-NER) |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Xeroderma Pigmentosum, Complementation Group D | 0.565428837 | 25 | 9 | BeFree_CLINVAR_CTD_human_MGD_ORPHANET_UNIPROT |
| Photosensitive Trichothiodystrophy | 0.44 | 6 | 6 | CLINVAR_CTD_human_MGD_UNIPROT |
| Cerebrooculofacioskeletal Syndrome 2 | 0.36 | 1 | 2 | CLINVAR_CTD_human_UNIPROT |
| Cerebrooculofacioskeletal Syndrome 1 | 0.240542884 | 2 | 2 | BeFree_CLINVAR_ORPHANET |
| Malignant neoplasm of lung | 0.213843535 | 78 | 18 | BeFree_GAD_GWASCAT |
| Squamous cell carcinoma | 0.175575822 | 23 | 2 | BeFree_CTD_human_GAD_LHGDN |
| Skin Neoplasms | 0.151486052 | 11 | 0 | CTD_human_GAD_LHGDN |
| Stomach Neoplasms | 0.144027638 | 9 | 0 | CTD_human_GAD_LHGDN |
| Colorectal Neoplasms | 0.142732561 | 9 | 0 | CTD_human_GAD_LHGDN |
| Trichothiodystrophy Syndromes | 0.137915164 | 66 | 2 | BeFree_ORPHANET |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。