CBL (Cbl proto-oncogene)
- symbol:
- CBL
- locus group:
- protein-coding gene
- location:
- 11q23.3
- gene_family:
- Ring finger proteins
- alias symbol:
- RNF55|c-Cbl
- alias name:
- oncogene CBL2
- entrez id:
- 867
- ensembl gene id:
- ENSG00000110395
- ucsc gene id:
- uc001pwe.6
- refseq accession:
- NM_005188
- hgnc_id:
- HGNC:1541
- approved reserved:
- 1989-06-30
CBL基因(Casitas B-lineage lymphoma proto-oncogene)属于CBL基因家族,该家族包括CBL、CBLB和CBLC三个成员,它们编码的蛋白质具有E3泛素连接酶活性,主要通过泛素化修饰(一种给蛋白质贴标签使其被降解的过程)调控细胞信号传导。CBL蛋白的主要功能是负向调节受体酪氨酸激酶(RTKs)和整合素等信号通路,通过标记这些受体使其被蛋白酶体降解,从而控制细胞生长、分化和迁移。CBL的作用位点集中在细胞膜和胞质内,与多种信号分子如EGFR、PDGFR等相互作用。当CBL发生功能丧失性突变时,会导致其泛素化功能缺陷,使得下游信号通路(如RAS-MAPK、PI3K-AKT)持续激活,促进细胞异常增殖,这与多种癌症(如白血病、肺癌)的发生相关。相反,CBL的过表达可能过度抑制生长信号,导致细胞功能异常。CBL基因家族成员的共性是含有高度保守的酪氨酸激酶结合域(TKB域)和RING指结构域(负责泛素连接酶活性),它们通过相似机制调控信号蛋白的稳定性。在免疫系统中,CBLB还参与T细胞耐受的调节,其缺失会导致自身免疫疾病。CBL的突变或表达异常也与努南综合征(Noonan syndrome)等发育障碍相关。此外,CBL的表达水平变化可能影响其他基因(如SRC、SYK)的活性,形成复杂的调控网络。在临床研究中,针对CBL突变肿瘤的靶向治疗(如蛋白酶体抑制剂)正在探索中。
This gene is a proto-oncogene that encodes a RING finger E3 ubiquitin ligase. The encoded protein is one of the enzymes required for targeting substrates for degradation by the proteasome. This protein mediates the transfer of ubiquitin from ubiquitin conjugating enzymes (E2) to specific substrates. This protein also contains an N-terminal phosphotyrosine binding domain that allows it to interact with numerous tyrosine-phosphorylated substrates and target them for proteasome degradation. As such it functions as a negative regulator of many signal transduction pathways. This gene has been found to be mutated or translocated in many cancers including acute myeloid leukaemia. Mutations in this gene are also the cause of Noonan syndrome-like disorder. [provided by RefSeq, Mar 2012]
该基因是一种原癌基因,其编码一个RING指的E3泛素连接酶。所编码的蛋白质是用于通过蛋白酶靶向底物降解所需的酶之一。该蛋白介导的泛素从泛素结合酶(E2),以特定的底物转移。该蛋白质还包含一个N-末端磷酸酪氨酸结合结构域,允许它与众多的酪氨??酸磷酸化底物相互作用和瞄准他们为蛋白酶体降解。因此,它用作许多信号转导途径的负调节剂。该基因已被发现被突变或在许多癌症中,包括急性髓性白血病易位。在这个基因的突变也努南综合征样病症的起因。 [由RefSeq的,2012年3月提供]
基因本体信息
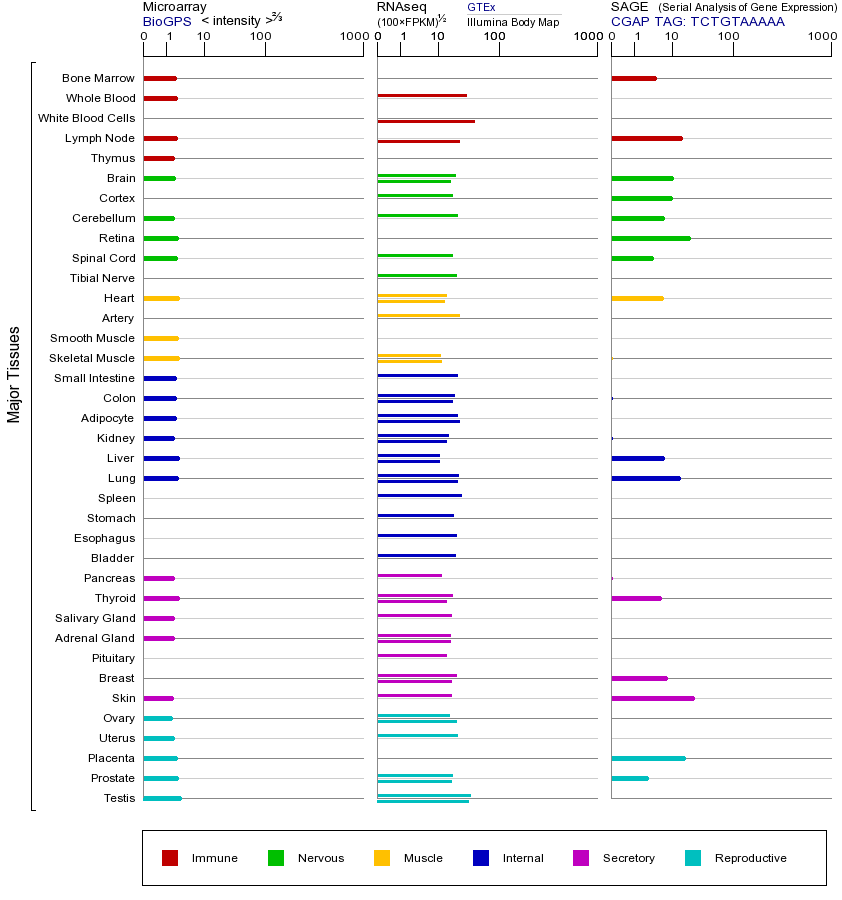
CBL基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
CBL基因的本体(GO)信息:
| 名称 |
|---|
| 4120 Ubiquitin mediated proteolysis [PATH:hsa04120] |
| 4012 ErbB signaling pathway [PATH:hsa04012] |
| 4630 Jak-STAT signaling pathway [PATH:hsa04630] |
| 4144 Endocytosis [PATH:hsa04144] |
| 4660 T cell receptor signaling pathway [PATH:hsa04660] |
| 4910 Insulin signaling pathway [PATH:hsa04910] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5205 Proteoglycans in cancer [PATH:hsa05205] |
| 5220 Chronic myeloid leukemia [PATH:hsa05220] |
| 5100 Bacterial invasion of epithelial cells [PATH:hsa05100] |
| 名称 |
|---|
| Adaptive Immune System |
| Antigen activates B Cell Receptor (BCR) leading to generation of second messengers |
| Constitutive Signaling by EGFRvIII |
| Constitutive Signaling by Ligand-Responsive EGFR Cancer Variants |
| Cytokine Signaling in Immune system |
| Disease |
| Diseases of signal transduction |
| EGFR downregulation |
| Immune System |
| Interleukin-3, 5 and GM-CSF signaling |
| Interleukin-6 signaling |
| Negative regulation of FGFR1 signaling |
| Negative regulation of FGFR2 signaling |
| Negative regulation of FGFR3 signaling |
| Negative regulation of FGFR4 signaling |
| Regulation of KIT signaling |
| Regulation of signaling by CBL |
| Signal Transduction |
| Signaling by EGFR |
| Signaling by EGFR in Cancer |
| Signaling by EGFRvIII in Cancer |
| Signaling by FGFR |
| Signaling by FGFR1 |
| Signaling by FGFR2 |
| Signaling by FGFR3 |
| Signaling by FGFR4 |
| Signaling by Interleukins |
| Signaling by Ligand-Responsive EGFR Variants in Cancer |
| Signaling by SCF-KIT |
| Signaling by TGF-beta Receptor Complex |
| Signaling by the B Cell Receptor (BCR) |
| Spry regulation of FGF signaling |
| TGF-beta receptor signaling activates SMADs |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| NOONAN SYNDROME-LIKE DISORDER WITH OR WITHOUT JUVENILE MYELOMONOCYTIC LEUKEMIA | 0.480271442 | 2 | 8 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Juvenile Myelomonocytic Leukemia | 0.364071628 | 17 | 2 | BeFree_CLINVAR_CTD_human_ORPHANET |
| Developmental Disabilities | 0.120271442 | 1 | 0 | BeFree_CTD_human |
| Cryptorchidism | 0.12 | 1 | 0 | CTD_human |
| Vasculitis | 0.12 | 1 | 0 | CTD_human |
| Growth Disorders | 0.12 | 1 | 0 | CTD_human |
| Leukemia, Myelocytic, Acute | 0.011073035 | 13 | 1 | BeFree_GAD_LHGDN |
| Leukemia, Myelomonocytic, Chronic | 0.003995683 | 7 | 0 | BeFree_GAD |
| Preleukemia | 0.002909916 | 3 | 0 | BeFree_GAD |
| Prostatic Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。