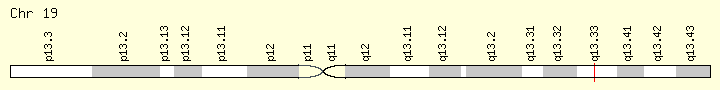BAX (BCL2 associated X, apoptosis regulator)
- symbol:
- BAX
- locus group:
- protein-coding gene
- location:
- 19q13.33
- gene_family:
- alias symbol:
- BCL2L4
- alias name:
- None
- entrez id:
- 581
- ensembl gene id:
- ENSG00000087088
- ucsc gene id:
- uc002plk.4
- refseq accession:
- NM_138763
- hgnc_id:
- HGNC:959
- approved reserved:
- 1994-11-08
BAX(Bcl-2-associated X protein)是Bcl-2基因家族的重要成员,属于促凋亡蛋白家族。Bcl-2家族主要调控细胞凋亡(程序性细胞死亡),其成员可分为抗凋亡蛋白(如Bcl-2、Bcl-xL)和促凋亡蛋白(如BAX、BAK)。BAX在正常情况下以单体形式存在于细胞质中,当受到凋亡信号(如DNA损伤或生长因子缺乏)刺激时,BAX会发生构象变化并转移到线粒体外膜,形成寡聚体,导致线粒体外膜通透性增加,释放细胞色素C等促凋亡因子,进而激活caspase级联反应,最终引发细胞凋亡。BAX的功能对维持组织稳态和清除异常细胞至关重要。BAX突变可能导致其促凋亡功能丧失,与多种癌症的发生发展相关,例如某些白血病和结直肠癌中BAX功能异常会导致细胞过度增殖。此外,BAX表达降低也会削弱细胞对化疗药物的敏感性。相反,BAX过表达会过度激活凋亡通路,可能导致组织损伤或退行性疾病,如神经退行性疾病。BAX与p53肿瘤 suppressor基因关系密切,p53可上调BAX表达以促进受损细胞的清除。BAX还与其他Bcl-2家族成员相互作用,其活性受到抗凋亡蛋白(如Bcl-2)的抑制,两者平衡决定了细胞命运。在癌症治疗中,靶向BAX或调节其活性是潜在策略,例如通过小分子药物激活BAX来增强肿瘤细胞对凋亡的敏感性。
The protein encoded by this gene belongs to the BCL2 protein family. BCL2 family members form hetero- or homodimers and act as anti- or pro-apoptotic regulators that are involved in a wide variety of cellular activities. This protein forms a heterodimer with BCL2, and functions as an apoptotic activator. This protein is reported to interact with, and increase the opening of, the mitochondrial voltage-dependent anion channel (VDAC), which leads to the loss in membrane potential and the release of cytochrome c. The expression of this gene is regulated by the tumor suppressor P53 and has been shown to be involved in P53-mediated apoptosis. Multiple alternatively spliced transcript variants, which encode different isoforms, have been reported for this gene. [provided by RefSeq, Jul 2008]
由该基因编码的蛋白质属于BCL2蛋白家族。 BCL2家族成员形成异源或同源二聚体和作用中所涉及的各种细胞活动,作为抗或促凋亡调节剂。此蛋白与BCL2的异二聚体,并且用作凋亡活化剂。这种蛋白质被报告进行交互,和增加的开口,线粒体电压依赖性阴离子通道(VDAC),这导致在膜电位丧失和细胞色素C的释放。该基因的表达是通过肿瘤抑制基因p53调节的,并已显示出参与了p53介导的细胞凋亡。多个可变剪接转录物变体,其编码不同同种型,已经报道了这个基因。 [由RefSeq的,2008年7月提供]
基因本体信息
BAX基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
BAX基因的本体(GO)信息:
| 名称 |
|---|
| 4141 Protein processing in endoplasmic reticulum [PATH:hsa04141] |
| 4071 Sphingolipid signaling pathway [PATH:hsa04071] |
| 4210 Apoptosis [PATH:hsa04210] |
| 4115 p53 signaling pathway [PATH:hsa04115] |
| 4722 Neurotrophin signaling pathway [PATH:hsa04722] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5203 Viral carcinogenesis [PATH:hsa05203] |
| 5210 Colorectal cancer [PATH:hsa05210] |
| 5014 Amyotrophic lateral sclerosis (ALS) [PATH:hsa05014] |
| 5016 Huntington's disease [PATH:hsa05016] |
| 5020 Prion diseases [PATH:hsa05020] |
| 4932 Non-alcoholic fatty liver disease (NAFLD) [PATH:hsa04932] |
| 5152 Tuberculosis [PATH:hsa05152] |
| 5166 HTLV-I infection [PATH:hsa05166] |
| 5161 Hepatitis B [PATH:hsa05161] |
| 名称 |
|---|
| Activation, translocation and oligomerization of BAX |
| Apoptosis |
| Intrinsic Pathway for Apoptosis |
| Programmed Cell Death |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Alzheimer's Disease | 0.2 | 4 | 0 | CTD_human_RGD |
| Acute kidney injury | 0.2 | 2 | 0 | CTD_human_RGD |
| Diabetes Mellitus, Experimental | 0.2 | 3 | 0 | CTD_human_RGD |
| Myocardial Reperfusion Injury | 0.2 | 2 | 0 | CTD_human_RGD |
| Mammary Neoplasms | 0.133893193 | 7 | 0 | BeFree_CTD_human_LHGDN |
| Colonic Neoplasms | 0.131711727 | 8 | 0 | BeFree_CTD_human_LHGDN |
| Prostatic Neoplasms | 0.1254487 | 3 | 0 | CTD_human_LHGDN |
| Esophageal Neoplasms | 0.12272435 | 1 | 0 | CTD_human_LHGDN |
| Colon Carcinoma | 0.122714419 | 10 | 2 | BeFree_CLINVAR |
| Myocardial Infarction | 0.120814326 | 5 | 0 | BeFree_CTD_human |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。